Science & Tech Spotlight: Genomic Sequencing Of Infectious Pathogens
Fast Facts
Genomic sequencing technology has the potential to improve how we monitor and treat infectious diseases. By revealing the genetic codes of pathogens, it can allow researchers to develop targeted vaccines, track new variants of the virus that causes COVID-19, and more.
Newer sequencing technologies are faster and more affordable. But widespread use for disease surveillance would require more laboratories to have infrastructure such as computer capacity, and trained personnel to work with the data.
Other challenges include high startup costs and privacy concerns over data that might be used to identify individuals who test positive for disease.

Highlights
Why this Matters
Genomic sequencing reveals the genetic code of an infectious disease pathogen, such as SARS-CoV-2, the virus that causes COVID-19. Newer, faster, and less costly sequencing can now be used to more quickly track transmission, detect new variants, and develop vaccines and other countermeasures. However, challenges such as high startup costs and privacy concerns remain.
The Technology
What is it? Genomic sequencing technologies decode a pathogen's genetic material by identifying the order of chemical "letters" of its DNA (or RNA, its chemical equivalent in some viruses). Each of four letters represents a chemical unit called a base. The sequence of the bases can reveal useful information for combatting disease. For example, sequencing of SARS-CoV-2 is used to track the spread of various strains, known as variants.
How does it work? Technologies for sequencing genomes of pathogens use chemicals to break up the DNA or RNA into small fragments. A first-generation method called Sanger sequencing repeatedly copies the fragments, fluorescently tags the copies, sorts them, and reads the letters of the genetic code. Sanger sequencing produces accurate data, but is slow because it reconstructs the genomic sequence base by base. This makes it expensive for large-scale sequencing of whole genomes.
Newer technologies such as next generation sequencing (NGS) can read much longer strings of letters from samples. One NGS technology works similarly to Sanger sequencing, but works in parallel on different parts of the genome at the same time, followed by computational reconstruction of the entire genome. Another type of NGS technology uses electrical current to thread long, single DNA strands through tiny pores of a membrane to identify the letters of the code. NGS can process millions to billions of sequences at the same time. Compared with Sanger sequencing, NGS reduces cost by over 1,000 times for larger samples. It also greatly reduces the time to determine the whole genome sequence of pathogens from multiple clinical samples, potentially allowing for more rapid discovery of variants.
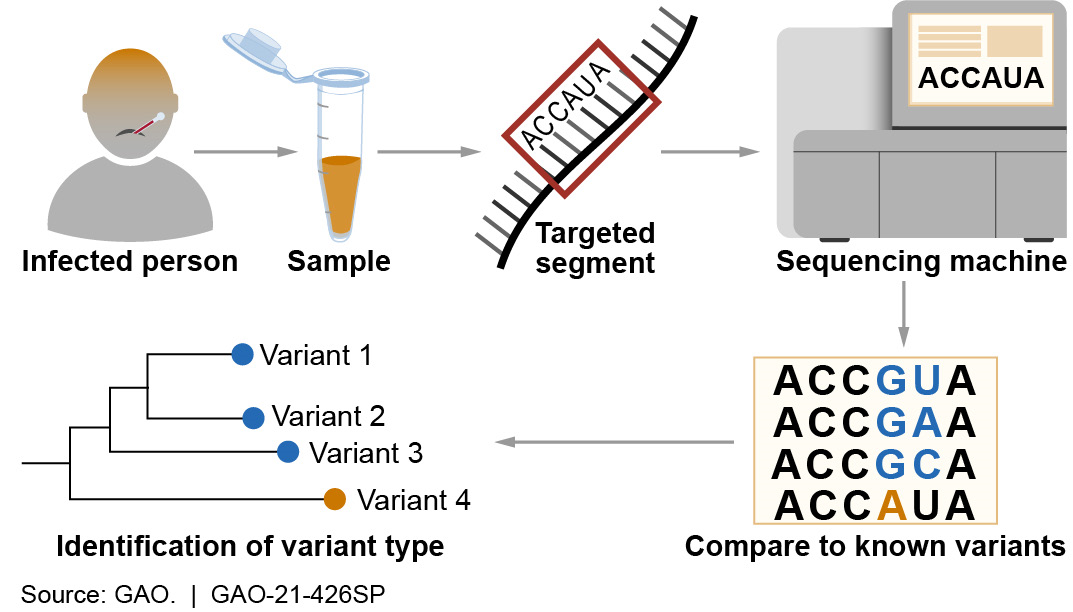
Figure 1. Use of genomic sequencing in the identification of infectious pathogen variants. A, C, G, and U represent letters of the genetic code.
How mature is it? First-generation sequencing is often used to confirm results of NGS. NGS is relatively new to the public health field, but is used to augment surveillance (i.e., data collection and analysis) for SARS-CoV-2 in the U.S. and overseas. New technologies are allowing greater access to sequencing capabilities by making NGS portable, faster, and more affordable (the cost of one sequence run is now one-millionth of what it was two decades ago). Genomic sequencing technologies enable many different areas of infectious disease study. For example, they enable genomic epidemiology—the science of using pathogen genomic data to determine the distribution and spread of an infectious disease in a group of people or animals, and the application of this information to respond to health problems.
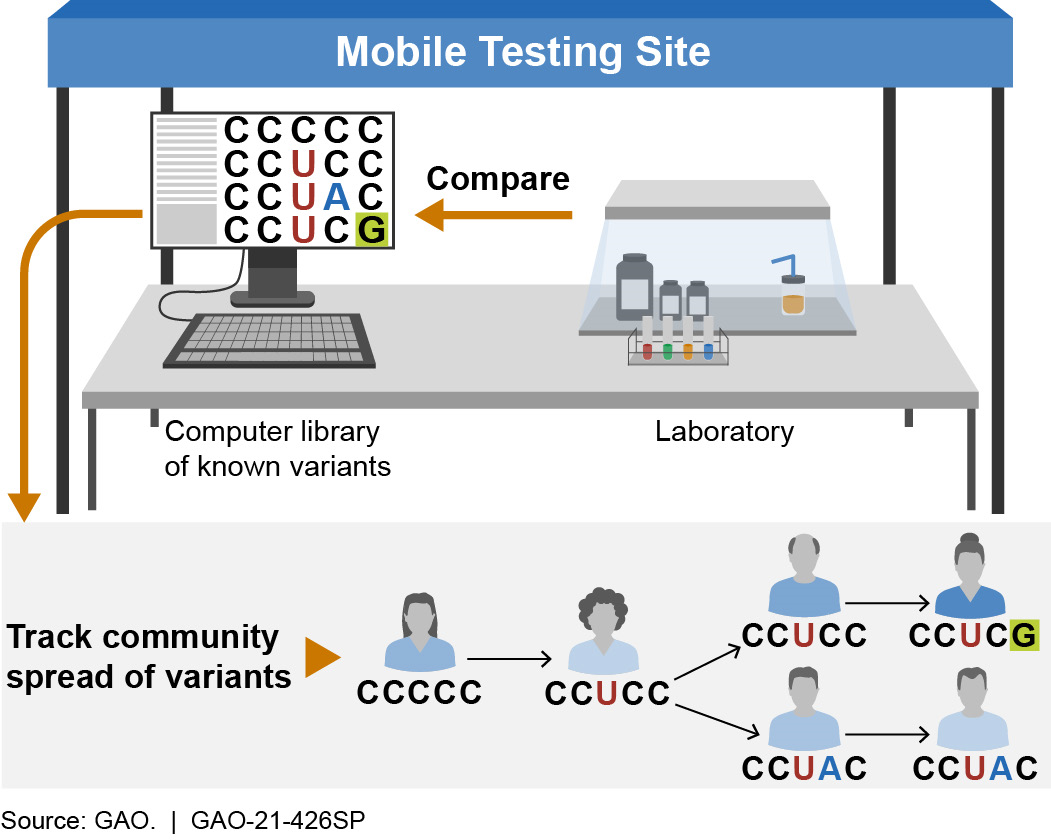
Figure 2. Use of mobile genomic sequencing technology to track the community spread of infectious pathogen variants. A, C, G, and U represent letters of the genetic code.
Wider use of NGS requires more laboratories to have infrastructure such as DNA extraction expertise, computer capacity and storage, and appropriately trained personnel to analyze and interpret sequencing data.
Opportunities
- Public health response. Genomic sequencing has the potential to transform public health approaches to infectious disease surveillance and treatment. It may allow identification of new pathogen variants shortly after they appear, generation of more data to estimate the prevalence of variants in populations, and eventually development of targeted treatments. For example, in February 2021, CDC announced a new investment of $200 million to identify, track, and mitigate emerging variants of SARS-CoV-2 through expansion of genomic sequencing. CDC stated that increasing sequencing of samples will improve the agency's ability to detect emerging variants and understand their spread with greater precision.
- Vaccine development. Analyses of data from large numbers of pathogens can serve as a basis to identify vaccine candidates and ensure that they are effective against common and emergent variants of a pathogen. For example, NGS was used in the development of the current messenger-RNA-based vaccines against SARS-CoV-2. If more samples of COVID-19 are tested for genomic variants, scientists may identify them faster, potentially enabling vaccine manufacturers to quickly modify vaccines to make them work better against such variants.
- Detection of emerging infectious diseases. Sequencing pathogens can help to detect diseases such as those transmitted between animals and humans before they emerge. Development of smaller, mobile, more affordable sequencing technology may make it more available for work in the community.
Challenges
There are a number of challenges to making genomic sequencing technologies more widely used.
- Up-front cost. Although the cost per sequence is far less than with first-generation sequencing, NGS requires significant up-front investment in laboratory equipment, computer resources, and training.
- Sensitivity. NGS requires that millions of copies of genetic material from the pathogen of interest are available. If there are too few in the sample, sequencing may not yield a useful result.
- Data sharing and privacy. Developing countermeasures for infectious disease outbreaks might require genomic data to be processed, shared, and linked in ways that could risk disclosure of the information or be used to potentially identify people who may have been infected with or transmitted new and deadly variants of a pathogen. If that information were to be made public, it might result in discrimination or stigmatization of those who test positive.
- National security concerns. Increased access to sequencing technologies could raise security concerns because state or non-state actors could use NGS in concert with gene editing technology to effectively modify pathogens’ survivability, virulence, and drug resistance as part of a biological warfare effort.
Policy Context & Questions
- What steps can policymakers take to prioritize creation of public health surveillance infrastructure (such as NGS) to build capabilities that help the nation more quickly detect and address infectious disease outbreaks?
- How can sponsors such as government, academic institutions, and private philanthropies leverage resources to support and broaden the use of genomic sequencing technologies, such as NGS, in the fight against COVID-19 and other infectious diseases?
- What additional safeguards could be considered to ensure confidentiality, security, and privacy in the context of genomic sequencing for public health?
For more information, contact Karen Howard at 202-512-6888 or HowardK@gao.gov.
